Prof. Bratati Kahali’s Lab at Centre for Brain Research (CBR) utilizes SahasraT to identify large population level genomic variations with at least thousands of human whole genome sequencing data. This has been achieved with the help of datastores that are efficient in processing the variants in a single process multiple data (SPMD) fashion which leads to increased performance in fetching the genomic data for tens of thousands of human samples.
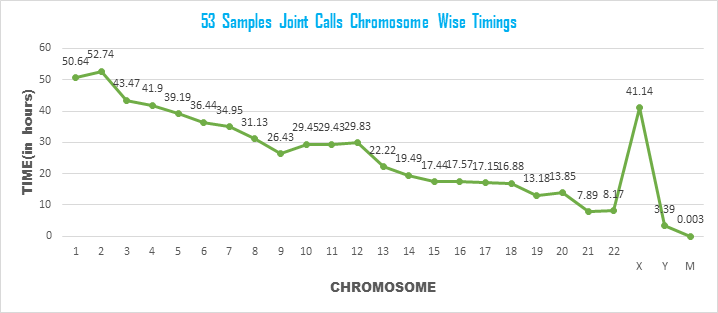
Experimental Setup: 600 cores of Sahasrat Cray XC40 were used for this experiment. The jobs were submitted in small queue upon 25 nodes. Total of 53 samples g.vcf files were consolidated into chromosome wise datastores using special genomics datastore utilities of GATK in 25 individual nodes of Cray. Upon the generated datastore using genotypeGVCF of GATK joint call is performed and final chromosome level VCF files are obtained. MPI based parallel codes in python were written which helped in processing 25 chromosomes [chromosome-1to22; X,Y,M] in parallel upon 25 compute nodes (each chromosome mapped to each node).
